RetrogeneDB ID: | retro_mmus_170 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 7:107209509..107210667(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000073894 | |
| Aliases: | Rbmxl2, 1700012H05Rik, Hnrnpg-t | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Rbmx | ||
| Ensembl ID: | ENSMUSG00000031134 | ||
| Aliases: | Rbmx, hnRNP G | ||
| Description: | RNA binding motif protein, X chromosome [Source:MGI Symbol;Acc:MGI:1343044] |

Retrocopy-Parental alignment summary:
>retro_mmus_170
ATGGTTGAAGCCGACCGTCCAGGGAAGCTCTTCATCGGCGGTCTCAACCTCGAGACCGACGAGAAGGGCCTCGAAACCGC
TTTCGGCAAGTATGGCCGCATCATCGAGGTGCTCCTGATGAAGGACCGGGAAACCTCCAAGTCCAGAGGCTTCGCCTTCG
TCACCTTCGAGAACCCCGCGGACGCCAAGGCCGCAGCCCGAGACATGAACGGCAAGTCCCTGGATGGTAAGGCCATCAAA
GTGGCCCAAGCCACCAAGCCGGTGTTCGAGAGCGGCCGGCGCGGGCCCCCTCCTCCCCGAAGCCGCGGCCGCCCGAGGGG
CCTGCGCGGGACCCGTGGCGGCGGCCCGAGGCGCCCTCCCTCCCGGGGCGGGTCGGCCGACGAAGGCGGCTACACGGGGG
ATTTCGACCTGCGGCCTTCGCGGGCCCCCATGCCACTGAAGCGCGGGCCGCCGCCGCGCCGGGCCGGGCCTCCGCCCAAA
AGAGCAGCGCCGTCGGGCCCCGCGCGCAGCGGCGCGGGAATTCGCGGGCGGGCGGCGGGCTCCCGGGGACGAGACGGCTA
CGGGGGCCCGCCGCGCCGGGAGCTGCCACCGCCGCGCCGCGACCCGTACCTGGGCCCGCGCGACGAGGGCTACTCGCCCC
GGGAGAGCTATTCGAGCCGCGACTACCCGAGCGCCCGGGACCCGCGCGACTTCGCGCCCTCGCCCCGCGACTACACCTAC
CGCGACTATGGCCACTCGAGCGCGCGAGACGAATGCCCTTCCCGGGGCTACTGTGAGCGCGACGGCTACGGCGGCCGCGA
GCGCGACTACGAGCATCCTAGCGGAGGCTCCTACCGCGACCCCTTCGACAGCTACGGGGACCCGCGTGGCGCCGGTCCTG
CCCGAGGGCCACCGCCATCTTACGGCGGGGGCCGCTATGACGAGTACCGAGGCTGCTCGCCCGACGGCTATGGCGGCCGC
GACAGCTACCGCAGCGAGCGCTACTCGAGTGGCCGTGAGCGCGTGGGTAGGCCGGAGCGCGGGCTGCCGCCGTCTGTAGA
ACGAAACTGCCCGGCGCCGCGGGATTCCTACAGCCGCTCGGGCCGCCGGGCGCCCCCGAGGGGCGGAGGCCGCGTGGGAA
GCCGCCTGGAGCGAGGGGGAGGCCGGAGCAGGTACTGA
ORF - retro_mmus_170 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 75.13 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MVEADRPGKLFIGGLNTETNEKALEAVFGKYGRIVEVLLMKDRETNKSRGFAFVTFESPADAKDAARDMN |
| MVEADRPGKLFIGGLN.ET.EK.LE..FGKYGRI.EVLLMKDRET.KSRGFAFVTFE.PADAK.AARDMN | |
| Retrocopy | MVEADRPGKLFIGGLNLETDEKGLETAFGKYGRIIEVLLMKDRETSKSRGFAFVTFENPADAKAAARDMN |
| Parental | GKSLDGKAIKVEQATKPSFESGRRGLPPPPRSRGPPRGLRGGRGGSGGTRGPPSRGGHMDDGGYSMNFTM |
| GKSLDGKAIKV.QATKP.FESGRRG.PPPPRSRG.PRGLRG.RGG..G.R.PPSRGG..D.GGY...F.. | |
| Retrocopy | GKSLDGKAIKVAQATKPVFESGRRG-PPPPRSRGRPRGLRGTRGG--GPRRPPSRGGSADEGGYTGDFDL |
| Parental | SSSRGPLPVKRGPPPRSGGPPPKRSAPSGPVRSSSGLGGRAPVSRGRDGYGGPPRREPLPSRRDVYLSPR |
| ..SR.P.P.KRGPPPR..GPPPKR.APSGP.RS..G..GRA..SRGRDGYGGPPRRE..P.RRD.YL.PR | |
| Retrocopy | RPSRAPMPLKRGPPPRRAGPPPKRAAPSGPARSGAGIRGRAAGSRGRDGYGGPPRRELPPPRRDPYLGPR |
| Parental | DDGYSTKDSYSSREYPSSRDTRDYAPPPRDYTYRDYGHSSSRDDYPSRGYSDRDGY-GRDRDYSDHPSGG |
| D.GYS...SYSSR.YPS.RD.RD.AP.PRDYTYRDYGHSS.RD..PSRGY..RDGY.GR.RDY..HPSGG | |
| Retrocopy | DEGYSPRESYSSRDYPSARDPRDFAPSPRDYTYRDYGHSSARDECPSRGYCERDGYGGRERDY-EHPSGG |
| Parental | SYRDSYESYGNSRSAPPTRGPPPSYGGSSRYDDY-SSSRDGYGGSRDSYSSSRSDLYSSGRDRVGRQERG |
| SYRD...SYG..R.A.P.RGPPPSYGG..RYD.Y...S.DGYGG.RDSY...RS..YSSGR.RVGR.ERG | |
| Retrocopy | SYRDPFDSYGDPRGAGPARGPPPSYGG-GRYDEYRGCSPDGYGG-RDSY---RSERYSSGRERVGRPERG |
| Parental | LPPSMERGYPPPRDSYSSSSRGA-PRGGGRGGSRSDRGGGRSRY |
| LPPS.ER..P.PRDSYS.S.R.A.PRGGGR.GSR..RGGGRSRY | |
| Retrocopy | LPPSVERNCPAPRDSYSRSGRRAPPRGGGRVGSRLERGGGRSRY |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 55 .88 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 61 .75 RPM |
| SRP007412_heart | 0 .00 RPM | 35 .52 RPM |
| SRP007412_kidney | 0 .00 RPM | 43 .47 RPM |
| SRP007412_liver | 0 .00 RPM | 14 .86 RPM |
| SRP007412_testis | 11 .48 RPM | 5 .44 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_170 was not detected
15 EST(s) were mapped to retro_mmus_170 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AV469074 | 107209477 | 107209729 | 99.7 | 251 | 1 | 250 |
| AV471529 | 107209462 | 107209813 | 99.2 | 347 | 3 | 343 |
| AV473722 | 107209460 | 107209811 | 99.8 | 349 | 1 | 347 |
| AV477899 | 107209459 | 107209813 | 100 | 352 | 0 | 350 |
| AV490614 | 107209486 | 107209813 | 100 | 325 | 0 | 324 |
| AV497252 | 107209459 | 107209813 | 100 | 353 | 0 | 352 |
| AV506265 | 107209486 | 107209813 | 100 | 324 | 0 | 322 |
| AV508702 | 107209463 | 107209813 | 99.8 | 340 | 0 | 337 |
| AV574611 | 107209455 | 107209813 | 100 | 357 | 0 | 356 |
| AV575020 | 107209460 | 107209813 | 99.8 | 351 | 1 | 349 |
| AV576406 | 107209472 | 107209813 | 99.8 | 338 | 1 | 335 |
| AV588165 | 107210025 | 107210299 | 99.3 | 271 | 2 | 268 |
| BB624528 | 107209464 | 107209990 | 97.2 | 516 | 10 | 504 |
| BG100571 | 107209420 | 107209959 | 97.6 | 526 | 11 | 512 |
| CF983176 | 107209421 | 107209980 | 98.1 | 548 | 11 | 537 |
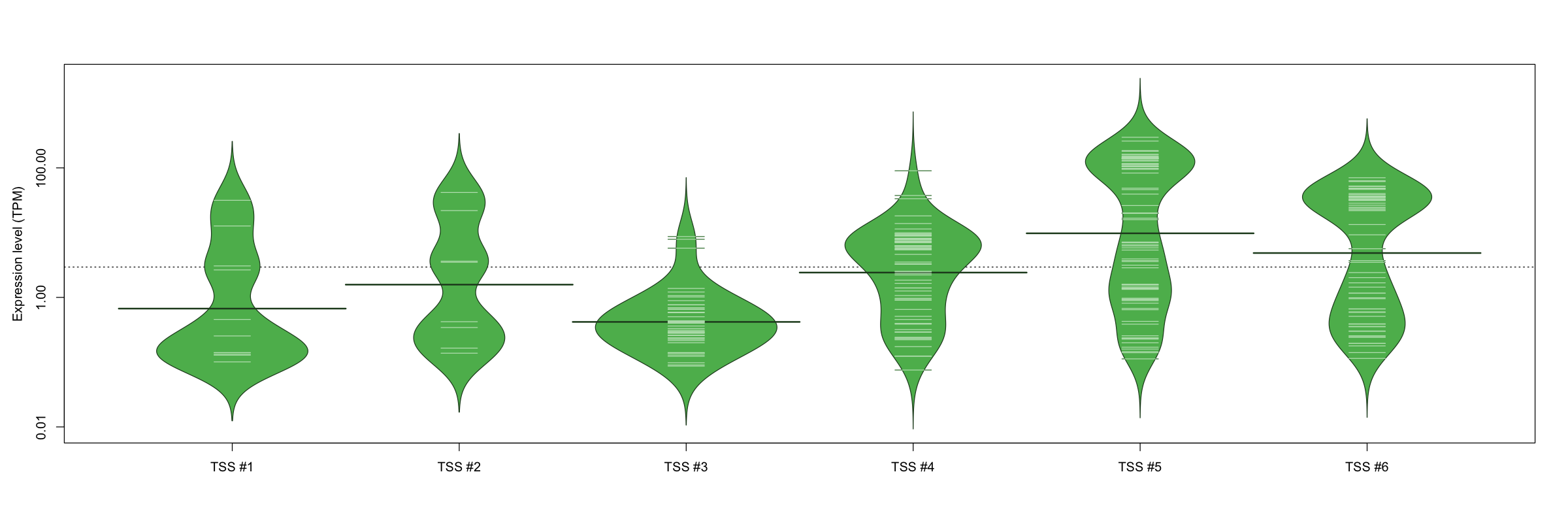
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_127499 | 1061 libraries | 7 libraries | 2 libraries | 0 libraries | 2 libraries |
| TSS #2 | TSS_127500 | 1064 libraries | 4 libraries | 2 libraries | 0 libraries | 2 libraries |
| TSS #3 | TSS_127502 | 1027 libraries | 38 libraries | 4 libraries | 3 libraries | 0 libraries |
| TSS #4 | TSS_127503 | 1006 libraries | 20 libraries | 14 libraries | 26 libraries | 6 libraries |
| TSS #5 | TSS_127504 | 996 libraries | 19 libraries | 13 libraries | 6 libraries | 38 libraries |
| TSS #6 | TSS_127505 | 1010 libraries | 21 libraries | 8 libraries | 2 libraries | 31 libraries |
The graphical summary, for retro_mmus_170 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_170 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_170 has 2 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Homo sapiens | retro_hsap_91 |
| Bos taurus | retro_btau_36 |