RetrogeneDB ID: | retro_mmus_3080 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 7:116824512..116825823(-) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Eef1g | ||
| Ensembl ID: | ENSMUSG00000071644 | ||
| Aliases: | Eef1g, 2610301D06Rik, AA407312, EF1G | ||
| Description: | eukaryotic translation elongation factor 1 gamma [Source:MGI Symbol;Acc:MGI:1914410] |

Retrocopy-Parental alignment summary:
>retro_mmus_3080
ATGGCGGCTGGGACCCTGTACACATACCCTGAAAATTGGAGGGCCTTCAAGGCCCTCATCGCTGCTCAGTACAGCGGGGC
TCAGGTCCGCGTGCTCTCCGCACCACCCCACTTCCACTTTGGCCAAACCAACCGCACCCCTGAATTTCTCCGAAAATTTC
CTGCCGGCAAGGTTCCAGCATTTGAGGGTGATGATGGATTCTGTGTGTTTGAGAGCAATGCCATTGCCTATTATGTAAGC
AATGAAGAGCTGCGAGGAAGTACGCCAGAGGCGGCAGCTCAGGTGGTGCAGTGGGTCAGCTTTGCTGACAGTGACATCGT
TCCTCCAGCTAGTACCTGGGTGTTCCCCACCTTGGGCATTATGCACCACAACAAACAGGCCACTGAAAATGCGAAAGAGG
AGGTGAAGCGAATCCTGGGGCTGCTGGACACTCACTTGAAGACTCGAACTTTTCTAGTGGGCGAGCGAGTGACACTGGCT
GATATTACAGTTGTCTGTACCCTGTTGTGGCTCTATAAACAGGTCTTAGAGCCTTCTTTTCGCCAGGCCTTCCCCAATAC
CAACCGGTGGTTCCTTACCTGCATTAACCAGCCCCAGTTCAGGGCTATCTTGGGGGAGGTGAAGCTGTGTGAGAAGATGG
CTCAGTTTGATGCTAAGAAGTTTGCAGAGAGCCAGTCTAAAAAAGATACCCCAAGGAAAGAAAAGGGTTCATGAGAGGAG
AAACAGAAACCCCAGGCTGAGCGGAAGGAGGAAAAAAATGCAGCTGCCCCAGCTCCTGAGGAAGAGATGGATGAGTGTGA
GCAGGCATTGGCTGCTGAGCCCAAGGCCAAGGACCCTTTCGCTCACCTGCCCAAGAGTACCTTTGTGTTGGATGAGTTTA
AGCGTAAGTACTCCAATGAGGACACCCTCTCTGTGGCACTGCCATATTTTTGGGAGCACTTTGATAAAGATGGCTGGTCC
CTGTGGTATGCCGAGTACCGCTTCCCTGAAGAGCTGACCCAGACCTTCATGAGTTGCAACCTCATCACTGGGATGTTTCA
GCGATTGGACAAACTGAGGAAGAATGCCTTTGCTAGTGTTATCCTCTTTGGAACCAACAACAGCAGCTCCATTTCTGGTG
TTTGGGTCTTCCGAGGCCAAGAGCTTGCCTTTCCGCTGAGTCCAGATTGGCAGGTGGACTATGAGTCGTATACATGGCGG
AAACTGGATCCTGGGAGCGAAGAAACCCAGACCCTGGTTCGAGAGTACTTTTCCTGGGAGGGGACCTTCCAGCATGTGGG
CAAAGCCGTCAATCAAGGCAAGATCTTCAAG
ORF - retro_mmus_3080 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 99.31 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAAGTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHFGQTNRTPEFLRKFPAGKVPAFEGDDGFCVF |
| MAAGTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHFGQTNRTPEFLRKFPAGKVPAFEGDDGFCVF | |
| Retrocopy | MAAGTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHFGQTNRTPEFLRKFPAGKVPAFEGDDGFCVF |
| Parental | ESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILG |
| ESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILG | |
| Retrocopy | ESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILG |
| Parental | LLDTHLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLC |
| LLDTHLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLC | |
| Retrocopy | LLDTHLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLC |
| Parental | EKMAQFDAKKFAESQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAKDPF |
| EKMAQFDAKKFAESQ.KKDTPRKEKGS.EEKQKPQAERKEEK.AAAPAPEEEMDECEQALAAEPKAKDPF | |
| Retrocopy | EKMAQFDAKKFAESQSKKDTPRKEKGS*EEKQKPQAERKEEKNAAAPAPEEEMDECEQALAAEPKAKDPF |
| Parental | AHLPKSTFVLDEFKRKYSNEDTLSVALPYFWEHFDKDGWSLWYAEYRFPEELTQTFMSCNLITGMFQRLD |
| AHLPKSTFVLDEFKRKYSNEDTLSVALPYFWEHFDKDGWSLWYAEYRFPEELTQTFMSCNLITGMFQRLD | |
| Retrocopy | AHLPKSTFVLDEFKRKYSNEDTLSVALPYFWEHFDKDGWSLWYAEYRFPEELTQTFMSCNLITGMFQRLD |
| Parental | KLRKNAFASVILFGTNNSSSISGVWVFRGQELAFPLSPDWQVDYESYTWRKLDPGSEETQTLVREYFSWE |
| KLRKNAFASVILFGTNNSSSISGVWVFRGQELAFPLSPDWQVDYESYTWRKLDPGSEETQTLVREYFSWE | |
| Retrocopy | KLRKNAFASVILFGTNNSSSISGVWVFRGQELAFPLSPDWQVDYESYTWRKLDPGSEETQTLVREYFSWE |
| Parental | GTFQHVGKAVNQGKIFK |
| GTFQHVGKAVNQGKIFK | |
| Retrocopy | GTFQHVGKAVNQGKIFK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .47 RPM | 74 .27 RPM |
| SRP007412_cerebellum | 0 .22 RPM | 52 .71 RPM |
| SRP007412_heart | 0 .31 RPM | 74 .28 RPM |
| SRP007412_kidney | 0 .42 RPM | 74 .94 RPM |
| SRP007412_liver | 0 .11 RPM | 53 .74 RPM |
| SRP007412_testis | 1 .19 RPM | 179 .11 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_3080 was not detected
No EST(s) were mapped for retro_mmus_3080 retrocopy.
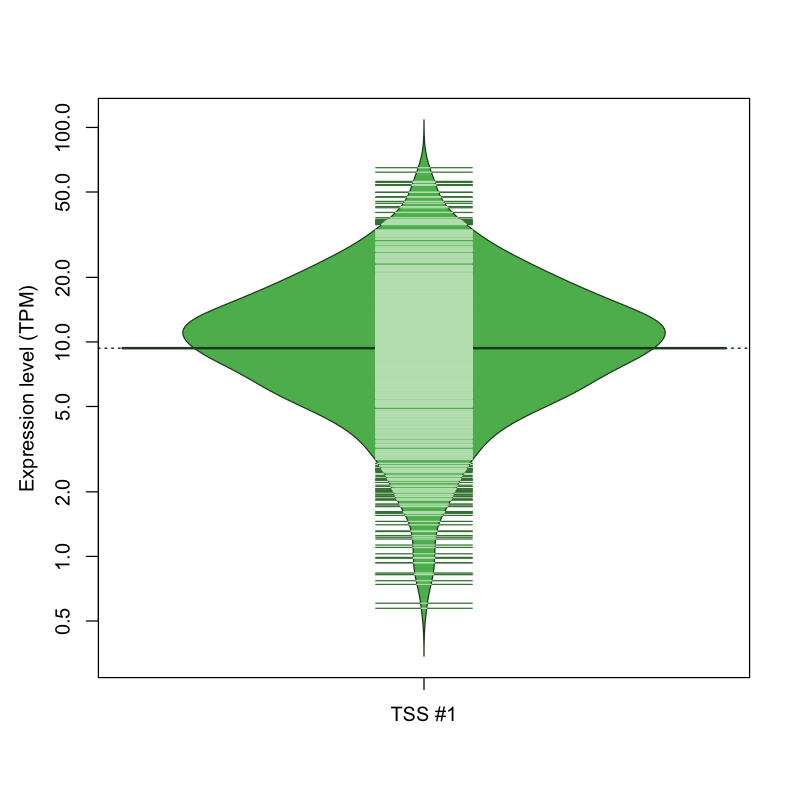
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_128041 | 51 libraries | 12 libraries | 162 libraries | 334 libraries | 513 libraries |
The graphical summary, for retro_mmus_3080 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3080 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3080 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 13 parental genes, and 53 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Canis familiaris | ENSCAFG00000015834 | 6 retrocopies | |
| Erinaceus europaeus | ENSEEUG00000001231 | 1 retrocopy | |
| Homo sapiens | ENSG00000254772 | 4 retrocopies | |
| Gorilla gorilla | ENSGGOG00000013557 | 12 retrocopies | |
| Latimeria chalumnae | ENSLACG00000014700 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000003339 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000006598 | 5 retrocopies | |
| Monodelphis domestica | ENSMODG00000007691 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000071644 | 7 retrocopies |
retro_mmus_1447, retro_mmus_1839, retro_mmus_2350, retro_mmus_2765, retro_mmus_3080 , retro_mmus_425, retro_mmus_989,
|
| Nomascus leucogenys | ENSNLEG00000003528 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000003768 | 3 retrocopies | |
| Rattus norvegicus | ENSRNOG00000020075 | 8 retrocopies | |
| Ictidomys tridecemlineatus | ENSSTOG00000002851 | 2 retrocopies |