RetrogeneDB ID: | retro_mmus_2887 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 7:3181092..3181885(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Psmb5 | ||
| Ensembl ID: | ENSMUSG00000022193 | ||
| Aliases: | None | ||
| Description: | proteasome (prosome, macropain) subunit, beta type 5 [Source:MGI Symbol;Acc:MGI:1194513] |

Retrocopy-Parental alignment summary:
>retro_mmus_2887
ATGGCACTGGCTAGCGTGTTGCAGCGGCCGATGCCGGTGAATCAGCACGGGTTTTTTGGGCTCGGAGGTGGTGCAGATCT
GCTGGACTTGGGTCCGGGGAGTCCTAGTGATGGGCTGAGCCTAGCCGCGCCGAGCTGGGGCGTCCCGGAGGAGCCGCGAA
TCGAAATGCTTCACGGAACCACCACCCTGGCCTTCAAGTTTCTCCATGGAGTCATTGTTGCAGCGGATTCCCGGGCCACA
GCAGGTGCTTATATTGCTTCCCAAGACGGTGAAGAAAGTAATAGAGATCAACCCGTATCTTCTGGGTACCATGGCTGGGG
GTGCAGCGGATTGCAGCTTCTGGGAGCGGTTGTTGGCTCGGCAGTGTCGAATCTATGAGCTTCGCAAAAAGGAACGCATC
TCGGTCGCAGCAGCCTCTAAACTGCTCGCTAACATGGTGTATCAGTACAAAGGCATGGGGCTGTCCATGGGCACCATGAT
CTGTGGCTGGGATAAGAGAGGCCCTGGCCTCTACTACGTAGACAGCGAGGGGAACAGGATCTCTGGGACCGCCTTCTCAG
TGGGCTCTGGCTCCGTGTATGCTTACGGCGTTATGGATCGAGGCTACTCCTATGACCTGAAAGTGGAGGAGGCCTATGAT
CTGGCCCGCCGAGCCATCTAACAAGCCACCTACAGAGATGCCTACTCCGGAGGGGCAGTCAACCTCTACCACGTGCGGGA
GGATGGCTGGATCCGGGTGTCCAGTGACAATGTAGCTGACTTACATGACAAGTACAGTAGTGTATCTGTCCCC
ORF - retro_mmus_2887 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 98.49 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected: | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | MALASVLQRPMPVNQHGFFGLGGGADLLDLGPGSPGDGLSLAAPSWGVPEEPRIEMLHGTTTLAFKFLHG |
| MALASVLQRPMPVNQHGFFGLGGGADLLDLGPGSP.DGLSLAAPSWGVPEEPRIEMLHGTTTLAFKFLHG | |
| Retrocopy | MALASVLQRPMPVNQHGFFGLGGGADLLDLGPGSPSDGLSLAAPSWGVPEEPRIEMLHGTTTLAFKFLHG |
| Parental | VIVAADSRATAGAYIASQ-TVKKVIEINPYLLGTMAGGAADCSFWERLLARQCRIYELRNKERISVAAAS |
| VIVAADSRATAGAYIASQ.TVKKVIEINPYLLGTMAGGAADCSFWERLLARQCRIYELR.KERISVAAAS | |
| Retrocopy | VIVAADSRATAGAYIASQ>TVKKVIEINPYLLGTMAGGAADCSFWERLLARQCRIYELRKKERISVAAAS |
| Parental | KLLANMVYQYKGMGLSMGTMICGWDKRGPGLYYVDSEGNRISGTAFSVGSGSVYAYGVMDRGYSYDLKVE |
| KLLANMVYQYKGMGLSMGTMICGWDKRGPGLYYVDSEGNRISGTAFSVGSGSVYAYGVMDRGYSYDLKVE | |
| Retrocopy | KLLANMVYQYKGMGLSMGTMICGWDKRGPGLYYVDSEGNRISGTAFSVGSGSVYAYGVMDRGYSYDLKVE |
| Parental | EAYDLARRAIYQATYRDAYSGGAVNLYHVREDGWIRVSSDNVADLHDKYSSVSVP |
| EAYDLARRAI.QATYRDAYSGGAVNLYHVREDGWIRVSSDNVADLHDKYSSVSVP | |
| Retrocopy | EAYDLARRAI*QATYRDAYSGGAVNLYHVREDGWIRVSSDNVADLHDKYSSVSVP |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 1 .75 RPM | 14 .02 RPM |
| SRP007412_cerebellum | 5 .64 RPM | 10 .20 RPM |
| SRP007412_heart | 0 .25 RPM | 25 .62 RPM |
| SRP007412_kidney | 7 .35 RPM | 16 .25 RPM |
| SRP007412_liver | 5 .52 RPM | 15 .10 RPM |
| SRP007412_testis | 7 .32 RPM | 14 .32 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_2887 was not detected
14 EST(s) were mapped to retro_mmus_2887 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AV448513 | 3181086 | 3181524 | 99.1 | 432 | 4 | 426 |
| AV482435 | 3181128 | 3181475 | 99.8 | 345 | 1 | 343 |
| AV483294 | 3181107 | 3181466 | 99.2 | 351 | 1 | 345 |
| AV511762 | 3181128 | 3181502 | 99.2 | 371 | 2 | 367 |
| BY037384 | 3181069 | 3181448 | 99.3 | 375 | 3 | 371 |
| BY039707 | 3181069 | 3181432 | 99.2 | 358 | 3 | 354 |
| BY045698 | 3181069 | 3181458 | 98.8 | 383 | 5 | 377 |
| BY047017 | 3181076 | 3181471 | 99.3 | 391 | 3 | 387 |
| BY049826 | 3181070 | 3181479 | 99.3 | 405 | 3 | 401 |
| BY051794 | 3181070 | 3181430 | 98.9 | 355 | 4 | 350 |
| BY058803 | 3181070 | 3181458 | 99.5 | 385 | 2 | 382 |
| BY303238 | 3181083 | 3181438 | 99.5 | 352 | 2 | 349 |
| BY314825 | 3181087 | 3181458 | 99.5 | 368 | 2 | 365 |
| BY339519 | 3181069 | 3181430 | 98.7 | 354 | 5 | 348 |
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_133977 | 271 libraries | 150 libraries | 97 libraries | 167 libraries | 387 libraries |
The graphical summary, for retro_mmus_2887 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
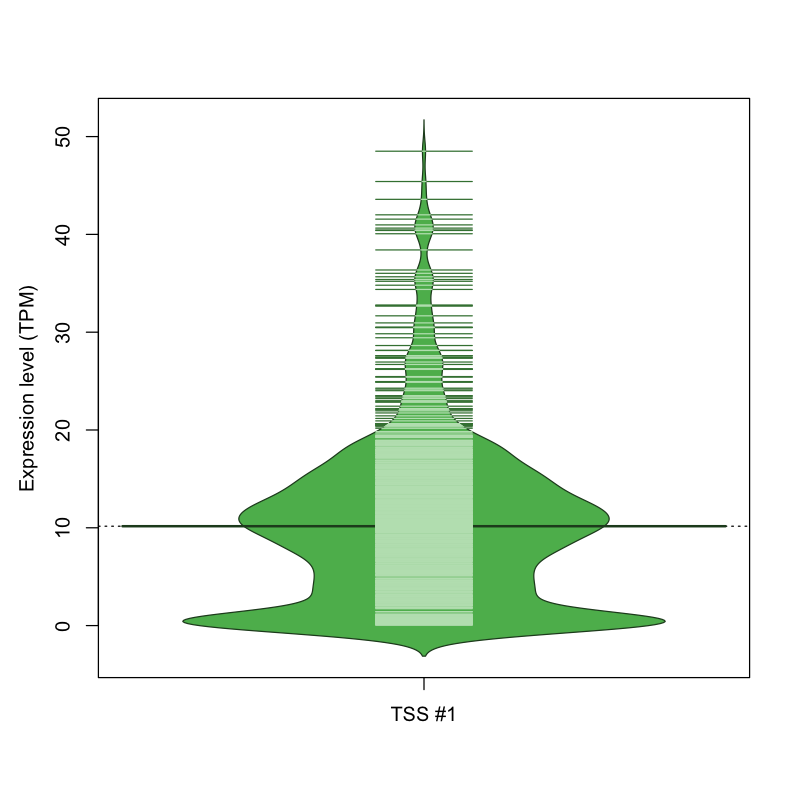
retro_mmus_2887 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2887 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 12 parental genes, and 18 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000008299 | 4 retrocopies | |
| Homo sapiens | ENSG00000100804 | 2 retrocopies | |
| Gorilla gorilla | ENSGGOG00000026856 | 2 retrocopies | |
| Latimeria chalumnae | ENSLACG00000015194 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000003572 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000014281 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000006007 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000022193 | 1 retrocopy |
retro_mmus_2887 ,
|
| Nomascus leucogenys | ENSNLEG00000014891 | 2 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000013109 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000005654 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000002036 | 1 retrocopy |