RetrogeneDB ID: | retro_mmus_1182 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 14:18531362..18532604(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000035129 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Fdft1 | ||
| Ensembl ID: | ENSMUSG00000021273 | ||
| Aliases: | Fdft1, SQS, SS | ||
| Description: | farnesyl diphosphate farnesyl transferase 1 [Source:MGI Symbol;Acc:MGI:102706] |

Retrocopy-Parental alignment summary:
>retro_mmus_1182
ATGGAGTTCGTCAAGTGTGTAGGCCACCCGGAGGAGTTCTACAACCTGCTGCGATTTCCGCATGGGAGGCCGGCGGAATT
TTATACCCAAAATGGACCAGGACTCAGCAGCAGCTTGAAGACCTGCTACAAGTATCTCAATCAGACCAGTCGCAGCTTTG
CCGCGGTTATCCAGGCGCTGGATGGGGACATACGGCACGCCATATGTGTGTTCTACCTGGTTCTCCGAGCCCTGGATACA
GTGGAGGATGACATGAGCATCAGTGTAGAGAAGAAGATCCCACTGCTGTGTAACTTCCACACTTTCCTCTATGACCCAGA
GTGGCGTTTCACTGAGAGTAAGGAGAAGGTCCGACAAGTGCTGGAGGACTTCCCCACGATCTCCCTGGAGTTTAGAAATT
TGGCTGAGAAATATCAAACAGTGATCGATGACATCTGCCACAGGATGGGGTGTGGGATGGCAGAATTTGTAGACAAGGAT
GTGACCTCCAAACAGGACTGGGACAAGTACTGCCACTACTTTGCTGGGCTGGTGGGAATTGACCTTTCTTGTCTATTCTC
TGCCTCAGAGTTTGAAGACCCCATAGTTGGTGAAGACATAGAGTGTGCCAACTCAATGGGTCTGTTCCTGCAGAAAACAA
ATATCATTCGTGATTATCTGGAAGATCAACAGGAAGGAAGTAAGTTTTGGCCTCGGGAGGTGTGGGGCAGATACATTAAG
TTGGAAGACTTTGCTAAGCCAGAGAACGTAGATGTGGCCCTGCAGTGCTTGAATGAACTCATAACCAACGCCCTACAGCA
CATCCCTGACATCCTCACCTATCTGTCAAGGCTCCGGAACCAGAGTGTGTTTAACTTCTGTGCTATTCCACAGGTAATGG
CCATTGCCACACTGGCTGCCTGTTACAATAACCAGCAGGTATTCAGAGGAATAGTGAAGATTCGGAAGGGGCAAGCAGTC
ACCCTCATGATGGATGCCACCAACATGCCTGCCATCAAAGCTATCATATACCAGTACATAGAAGAGATTTATCACTGGAT
CCCCAACTCAGACCCATCATCAAGCAAAACCAAGCAGGTCATCTCCAACATCAGGACACAGAACCTTCCCAACTGCCAGC
TCATCTCCTGAAGCCACTACTCCCCCATTTACCTGTCATTTATCATGCTCTTGGCTGCCCTGAGCTGGCAGTACCTGAGC
ACCCTGTCCCAGGTCACAGAAGACTATGTCCAGAGAGAACAC
ORF - retro_mmus_1182 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 92.55 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MEFVKCLGHPEEFYNLLRFRMGGRRNFIPKMDQDSLSSSLKTCYKYLNQTSRSFAAVIQALDGDIRHAIC |
| MEFVKC.GHPEEFYNLLRF..G....F........LSSSLKTCYKYLNQTSRSFAAVIQALDGDIRHAIC | |
| Retrocopy | MEFVKCVGHPEEFYNLLRFPHGRPAEFYTQ-NGPGLSSSLKTCYKYLNQTSRSFAAVIQALDGDIRHAIC |
| Parental | VFYLVLRALDTVEDDMSISVEKKIPLLCNFHTFLYDPEWRFTESKEKDRQVLEDFPTISLEFRNLAEKYQ |
| VFYLVLRALDTVEDDMSISVEKKIPLLCNFHTFLYDPEWRFTESKEK.RQVLEDFPTISLEFRNLAEKYQ | |
| Retrocopy | VFYLVLRALDTVEDDMSISVEKKIPLLCNFHTFLYDPEWRFTESKEKVRQVLEDFPTISLEFRNLAEKYQ |
| Parental | TVIDDICHRMGCGMAEFVDKDVTSKQDWDKYCHYVAGLVGIGLSRLFSASEFEDPIVGEDIECANSMGLF |
| TVIDDICHRMGCGMAEFVDKDVTSKQDWDKYCHY.AGLVGI.LS.LFSASEFEDPIVGEDIECANSMGLF | |
| Retrocopy | TVIDDICHRMGCGMAEFVDKDVTSKQDWDKYCHYFAGLVGIDLSCLFSASEFEDPIVGEDIECANSMGLF |
| Parental | LQKTNIIRDYLEDQQEGRKFWPQEVWGRYIKKLEDFAKPENVDVAVQCLNELITNTLQHIPDVLTYLSRL |
| LQKTNIIRDYLEDQQEG.KFWP.EVWGRYI.KLEDFAKPENVDVA.QCLNELITN.LQHIPD.LTYLSRL | |
| Retrocopy | LQKTNIIRDYLEDQQEGSKFWPREVWGRYI-KLEDFAKPENVDVALQCLNELITNALQHIPDILTYLSRL |
| Parental | RNQSVFNFCAIPQVMAIATLAACYNNQQVFKGVVKIRKGQAVTLMMDATNMPAVKAIIYQYIEEIYHRIP |
| RNQSVFNFCAIPQVMAIATLAACYNNQQVF.G.VKIRKGQAVTLMMDATNMPA.KAIIYQYIEEIYH.IP | |
| Retrocopy | RNQSVFNFCAIPQVMAIATLAACYNNQQVFRGIVKIRKGQAVTLMMDATNMPAIKAIIYQYIEEIYHWIP |
| Parental | NSDPSSSKTKQVISKIRTQNLPNCQLISRSHYSPIYLSFIMLLAALSWQYLSTLSQVTEDYVQREH |
| NSDPSSSKTKQVIS.IRTQNLPNCQLIS.SHYSPIYLSFIMLLAALSWQYLSTLSQVTEDYVQREH | |
| Retrocopy | NSDPSSSKTKQVISNIRTQNLPNCQLIS*SHYSPIYLSFIMLLAALSWQYLSTLSQVTEDYVQREH |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .21 RPM | 52 .38 RPM |
| SRP007412_cerebellum | 0 .04 RPM | 29 .66 RPM |
| SRP007412_heart | 0 .09 RPM | 26 .46 RPM |
| SRP007412_kidney | 0 .06 RPM | 21 .68 RPM |
| SRP007412_liver | 0 .38 RPM | 137 .47 RPM |
| SRP007412_testis | 0 .32 RPM | 235 .69 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_1182 was not detected
No EST(s) were mapped for retro_mmus_1182 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_33071 | 762 libraries | 214 libraries | 94 libraries | 1 library | 1 library |
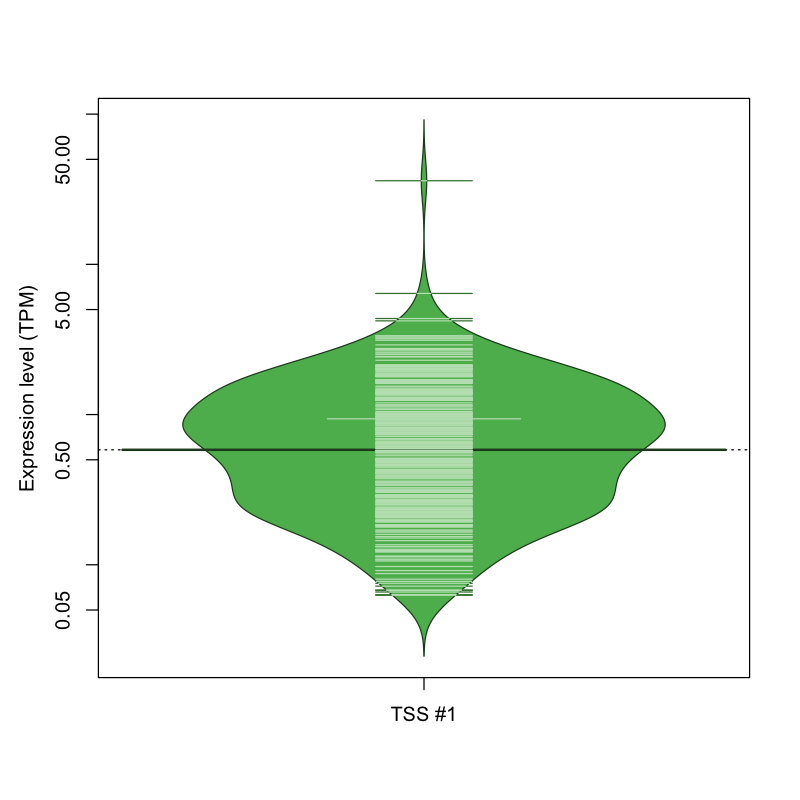
The graphical summary, for retro_mmus_1182 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1182 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1182 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 5 parental genes, and 6 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000003988 | 2 retrocopies | |
| Erinaceus europaeus | ENSEEUG00000013655 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000007372 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000021273 | 1 retrocopy |
retro_mmus_1182 ,
|
| Vicugna pacos | ENSVPAG00000006183 | 1 retrocopy |