RetrogeneDB ID: | retro_mmus_2944 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 7:72672817..72674275(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000097530 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Kansl2 | ||
| Ensembl ID: | ENSMUSG00000022992 | ||
| Aliases: | Kansl2, 2310037I24Rik, B930009D17 | ||
| Description: | KAT8 regulatory NSL complex subunit 2 [Source:MGI Symbol;Acc:MGI:1916862] |

Retrocopy-Parental alignment summary:
>retro_mmus_2944
ATGAACAGGATTCGGATTCACGTCTTGCCAACGAATCGGGGGAGGATCACCCCAGTGCCCAGGTCGCAGGAGCCCCTTTC
CTGTTCGTTCACTCACCGCCCGTGCTCTCAGCCGCGGCTGGAGGGGCAGGAGTTCTGCATTAAGCACATTCTTGAAGACA
AGAATGCACCCTTCAAACAATGTAGTTACGTCTCGACGAAGAATGGAAAAAGGTGCCCCAGTGCTGCCCCAAAGCCCGAG
AAGAAAGATGGGGTGTCTTTCTGTGCCGAACATGCTCGAAGAAATGCCCTAGCACTTCATGCTCAAATGAAGAAGAGCAA
CCCAGGCCCTATGGGTGAAACCCTCTTGTGCCAGTTGAGCTCATACGCTAAAACAGAGCTGGGCTCGCAGACCCCAGAGA
GCAGCCGCAGTGAAGCCAGCCGGATACTGGATGAAGACAGCTGGAGTGATGGGGACCAGGAACCCATCACTGTGGATCAG
ACATGGAGAGGTGACCCTGACAGTGAAGCCGATAGCATAGACAGTGATCAAGAAGATCCCCTGAAACATGCTGGTGTCTA
TACAGCTGAAGAAGTGGCTCTTATTATGAGAGAAAAGCTAATTCGTTTGCAGTCTTTGTACATTGATCAGTTTAAAAGAC
TCCAACACCTGCTCAAAGAGAAGAAGCGCCGCTATTTACACAACCGGAAAGTGGAGCACGAGGCTTTAGGTAGTAGTCTC
CTGACTGGCCCTGAGGGACTTTTGGCCAAAGAGAGAGAGAATTTAAAGCGACTAAAATGTCTTAGGCGATACCGTCAACG
CTATGGAGTAGAAGCCTTACTACACAGGCAGCTGAAGGAACGCAGAATGCTGGCCACCGATGGGGCTGCTCAACAGGCTC
ACACCACTCGTTCCAGTCAGAGGTGCTTAGCCTTTGTGGATGATGTGCGCTGCTCCAATCAGTCTCTCCCAATGACCAGA
CACTGCCTTACCCATATCTGTCAGGATACAAATCAGGTTCTCTTCAAATGCTGCCAGGGATCTGAAGAGGTCCCCTGCAA
CAAACCAGTTCCTGTAAGCCTTTCTGAGGATCCCTGCTGCCCTCTGCATTTCCAGTTGCCTCCTCAGATGTATAAGCCCG
AGTAGGTACTGTCTGTGCCAGACGATCTGGAAGCCGGCCCCATGGATCTGTACTTGAGTGCTGCCGAGCTTCAGCCCACT
GAGAGTTTACCACTGGAGTTCAGTGATGACTTGGATGTTGTGGGTGATGGCATGCCATGCCCGCCGTCGCCGTTGCTCTT
TGATCCTTCGCTAACTCTTGAAGATCATTCAGTCACAGAAAGTGCCGGAGACGCAGGACAGATGCAGGCGGCTGGGGATG
GGTGCAGATCCCAGGGAGCTCGAAGTGTAGAGAAAGCAGCATTCCCTCAGCGTGGATTGGCTACTGCGAATGGAAAACCA
GAACCCACTTCTATCAGT
ORF - retro_mmus_2944 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 99.79 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MNRIRIHVLPTNRGRITPVPRSQEPLSCSFTHRPCSQPRLEGQEFCIKHILEDKNAPFKQCSYVSTKNGK |
| MNRIRIHVLPTNRGRITPVPRSQEPLSCSFTHRPCSQPRLEGQEFCIKHILEDKNAPFKQCSYVSTKNGK | |
| Retrocopy | MNRIRIHVLPTNRGRITPVPRSQEPLSCSFTHRPCSQPRLEGQEFCIKHILEDKNAPFKQCSYVSTKNGK |
| Parental | RCPSAAPKPEKKDGVSFCAEHARRNALALHAQMKKSNPGPMGETLLCQLSSYAKTELGSQTPESSRSEAS |
| RCPSAAPKPEKKDGVSFCAEHARRNALALHAQMKKSNPGPMGETLLCQLSSYAKTELGSQTPESSRSEAS | |
| Retrocopy | RCPSAAPKPEKKDGVSFCAEHARRNALALHAQMKKSNPGPMGETLLCQLSSYAKTELGSQTPESSRSEAS |
| Parental | RILDEDSWSDGDQEPITVDQTWRGDPDSEADSIDSDQEDPLKHAGVYTAEEVALIMREKLIRLQSLYIDQ |
| RILDEDSWSDGDQEPITVDQTWRGDPDSEADSIDSDQEDPLKHAGVYTAEEVALIMREKLIRLQSLYIDQ | |
| Retrocopy | RILDEDSWSDGDQEPITVDQTWRGDPDSEADSIDSDQEDPLKHAGVYTAEEVALIMREKLIRLQSLYIDQ |
| Parental | FKRLQHLLKEKKRRYLHNRKVEHEALGSSLLTGPEGLLAKERENLKRLKCLRRYRQRYGVEALLHRQLKE |
| FKRLQHLLKEKKRRYLHNRKVEHEALGSSLLTGPEGLLAKERENLKRLKCLRRYRQRYGVEALLHRQLKE | |
| Retrocopy | FKRLQHLLKEKKRRYLHNRKVEHEALGSSLLTGPEGLLAKERENLKRLKCLRRYRQRYGVEALLHRQLKE |
| Parental | RRMLATDGAAQQAHTTRSSQRCLAFVDDVRCSNQSLPMTRHCLTHICQDTNQVLFKCCQGSEEVPCNKPV |
| RRMLATDGAAQQAHTTRSSQRCLAFVDDVRCSNQSLPMTRHCLTHICQDTNQVLFKCCQGSEEVPCNKPV | |
| Retrocopy | RRMLATDGAAQQAHTTRSSQRCLAFVDDVRCSNQSLPMTRHCLTHICQDTNQVLFKCCQGSEEVPCNKPV |
| Parental | PVSLSEDPCCPLHFQLPPQMYKPEQVLSVPDDLEAGPMDLYLSAAELQPTESLPLEFSDDLDVVGDGMPC |
| PVSLSEDPCCPLHFQLPPQMYKPE.VLSVPDDLEAGPMDLYLSAAELQPTESLPLEFSDDLDVVGDGMPC | |
| Retrocopy | PVSLSEDPCCPLHFQLPPQMYKPE*VLSVPDDLEAGPMDLYLSAAELQPTESLPLEFSDDLDVVGDGMPC |
| Parental | PPSPLLFDPSLTLEDHSVTESAGDAGQMQAAGDGCRSQGARSVEKAAFPQRGLATANGKPEPTSIS |
| PPSPLLFDPSLTLEDHSVTESAGDAGQMQAAGDGCRSQGARSVEKAAFPQRGLATANGKPEPTSIS | |
| Retrocopy | PPSPLLFDPSLTLEDHSVTESAGDAGQMQAAGDGCRSQGARSVEKAAFPQRGLATANGKPEPTSIS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .37 RPM | 8 .48 RPM |
| SRP007412_cerebellum | 1 .35 RPM | 8 .16 RPM |
| SRP007412_heart | 0 .16 RPM | 12 .95 RPM |
| SRP007412_kidney | 1 .12 RPM | 8 .39 RPM |
| SRP007412_liver | 0 .27 RPM | 5 .73 RPM |
| SRP007412_testis | 1 .19 RPM | 6 .91 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_2944 was not detected
28 EST(s) were mapped to retro_mmus_2944 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AI526657 | 72673357 | 72673503 | 100 | 146 | 0 | 146 |
| AV137827 | 72672972 | 72673172 | 95.5 | 191 | 9 | 182 |
| AV489273 | 72673401 | 72673886 | 100 | 482 | 0 | 479 |
| AV497846 | 72672808 | 72673313 | 100 | 503 | 0 | 502 |
| BE281765 | 72673034 | 72673720 | 99.2 | 570 | 1 | 564 |
| BE554249 | 72673045 | 72673267 | 100 | 221 | 0 | 221 |
| BE914316 | 72672814 | 72673622 | 99.9 | 678 | 0 | 665 |
| BG080856 | 72673059 | 72673697 | 100 | 638 | 0 | 638 |
| BQ572802 | 72672870 | 72673541 | 100 | 669 | 0 | 669 |
| BU612541 | 72672791 | 72673550 | 99.8 | 756 | 1 | 753 |
| BY136485 | 72672782 | 72673150 | 100 | 368 | 0 | 368 |
| BY698237 | 72673475 | 72673877 | 100 | 402 | 0 | 402 |
| CA531810 | 72672945 | 72673520 | 99.8 | 459 | 1 | 457 |
| CA531872 | 72672945 | 72673485 | 99.8 | 424 | 1 | 422 |
| CA554191 | 72673159 | 72673293 | 100 | 134 | 0 | 134 |
| CA566171 | 72672934 | 72673220 | 100 | 286 | 0 | 286 |
| CA566845 | 72672822 | 72673307 | 100 | 485 | 0 | 485 |
| CF154388 | 72673159 | 72673684 | 100 | 525 | 0 | 525 |
| CF157376 | 72673159 | 72673660 | 100 | 501 | 0 | 501 |
| CF724626 | 72673074 | 72673719 | 99.9 | 643 | 1 | 641 |
| CF724708 | 72673021 | 72673687 | 99.9 | 659 | 1 | 654 |
| CF892909 | 72672967 | 72673590 | 99.9 | 622 | 1 | 621 |
| CF893589 | 72672967 | 72673563 | 99.9 | 595 | 1 | 594 |
| CF897240 | 72672803 | 72673405 | 100 | 602 | 0 | 602 |
| CJ058625 | 72672786 | 72673223 | 99.8 | 435 | 1 | 434 |
| CJ069080 | 72672903 | 72673384 | 99.8 | 356 | 0 | 354 |
| CK391930 | 72672809 | 72673363 | 99.9 | 553 | 1 | 552 |
| CO425818 | 72672837 | 72673465 | 99.9 | 625 | 1 | 624 |
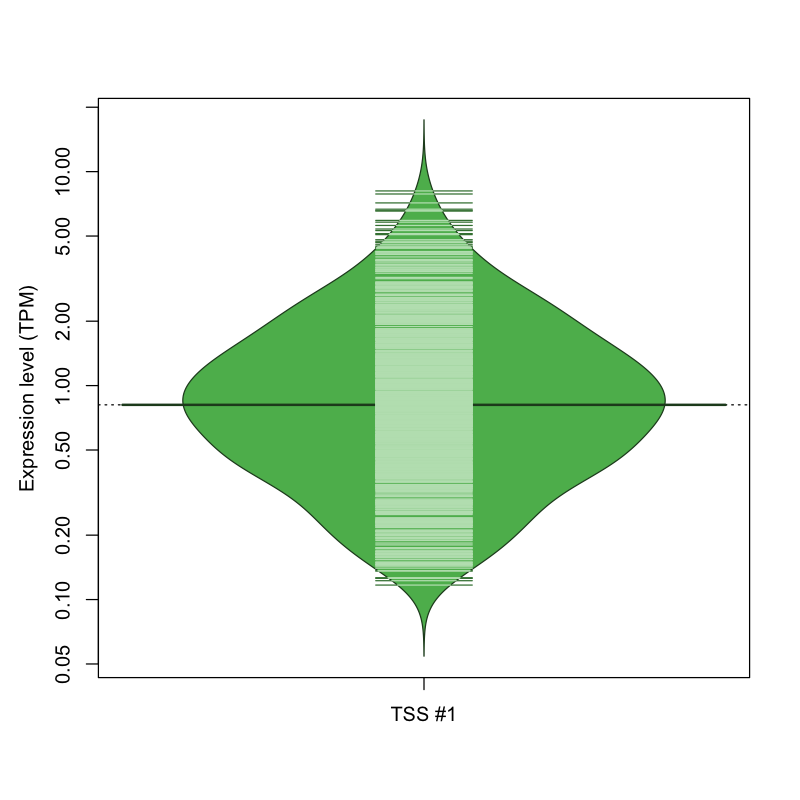
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_136885 | 271 libraries | 475 libraries | 313 libraries | 13 libraries | 0 libraries |
The graphical summary, for retro_mmus_2944 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2944 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2944 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 5 parental genes, and 8 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Cavia porcellus | ENSCPOG00000005760 | 1 retrocopy | |
| Dipodomys ordii | ENSDORG00000001180 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000022992 | 2 retrocopies |
retro_mmus_2944 , retro_mmus_3134,
|
| Rattus norvegicus | ENSRNOG00000010904 | 3 retrocopies | |
| Tupaia belangeri | ENSTBEG00000016536 | 1 retrocopy |