RetrogeneDB ID: | retro_mmus_1172 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 14:9886175..9886631(+) | ||
| Located in intron of: | ENSMUSG00000060579 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000089686 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Mnd1 | ||
| Ensembl ID: | ENSMUSG00000033752 | ||
| Aliases: | Mnd1, 2610034E18Rik, AI591601, Gaj | ||
| Description: | meiotic nuclear divisions 1 homolog (S. cerevisiae) [Source:MGI Symbol;Acc:MGI:1924165] |

Retrocopy-Parental alignment summary:
>retro_mmus_1172
CTAGTGGATGACGGTATGGTTGACTGCGAGAGGATCGGGACGTCCAATTACTACTGGGCTTTTCCGAGCAAGGCTCTTCA
TGCCAGGAAGCACAAGTTGGAGGCTCTGAACTCTCAGCTGTCAGAGGGAAGCCAGAAGCATGCAGACTTACAGAAGAGCA
TTGAGAAAGCAAGAGTTGGCCGGCAAGAGACGCAAGAACGAACCATGTTTGCAAAAGAACTTTCTTCATTTCGAGACTAA
AGGCAACAACTTAAGGCAGAAGTGGAAAAATATCGAGAATGTGACCCACAGGTTGTGGAAGAGATTCGTGAAGCAAATAA
AGTAGCTAAAGAAGCCGCCAATCGATGGACTGATAACATATTTGCAATAAAATCCTGGGCCAAAAGAAAATTTGGGTTTG
AAGAAAGTAAAATTGATAAAAACTTTGGAATTCCAGAAGACTTTGACTACATAGAC
ORF - retro_mmus_1172 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 96.71 % |
| Parental protein coverage: | 74.15 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | LVDDGMVDCERIGTSNYYWAFPSKALHARKRKLEALNSQLSEGSQKHADLQKSIEKARVGRQETEERAML |
| LVDDGMVDCERIGTSNYYWAFPSKALHARK.KLEALNSQLSEGSQKHADLQKSIEKARVGRQET.ER.M. | |
| Retrocopy | LVDDGMVDCERIGTSNYYWAFPSKALHARKHKLEALNSQLSEGSQKHADLQKSIEKARVGRQETQERTMF |
| Parental | AKELSSFRDQRQQLKAEVEKYRECDPQVVEEIREANKVAKEAANRWTDNIFAIKSWAKRKFGFEESKIDK |
| AKELSSFRD.RQQLKAEVEKYRECDPQVVEEIREANKVAKEAANRWTDNIFAIKSWAKRKFGFEESKIDK | |
| Retrocopy | AKELSSFRD*RQQLKAEVEKYRECDPQVVEEIREANKVAKEAANRWTDNIFAIKSWAKRKFGFEESKIDK |
| Parental | NFGIPEDFDYID |
| NFGIPEDFDYID | |
| Retrocopy | NFGIPEDFDYID |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 0 .14 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 0 .17 RPM |
| SRP007412_heart | 0 .00 RPM | 0 .72 RPM |
| SRP007412_kidney | 0 .00 RPM | 0 .60 RPM |
| SRP007412_liver | 0 .00 RPM | 2 .27 RPM |
| SRP007412_testis | 0 .00 RPM | 5 .40 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_1172 was not detected
No EST(s) were mapped for retro_mmus_1172 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_32395 | 748 libraries | 222 libraries | 92 libraries | 8 libraries | 2 libraries |
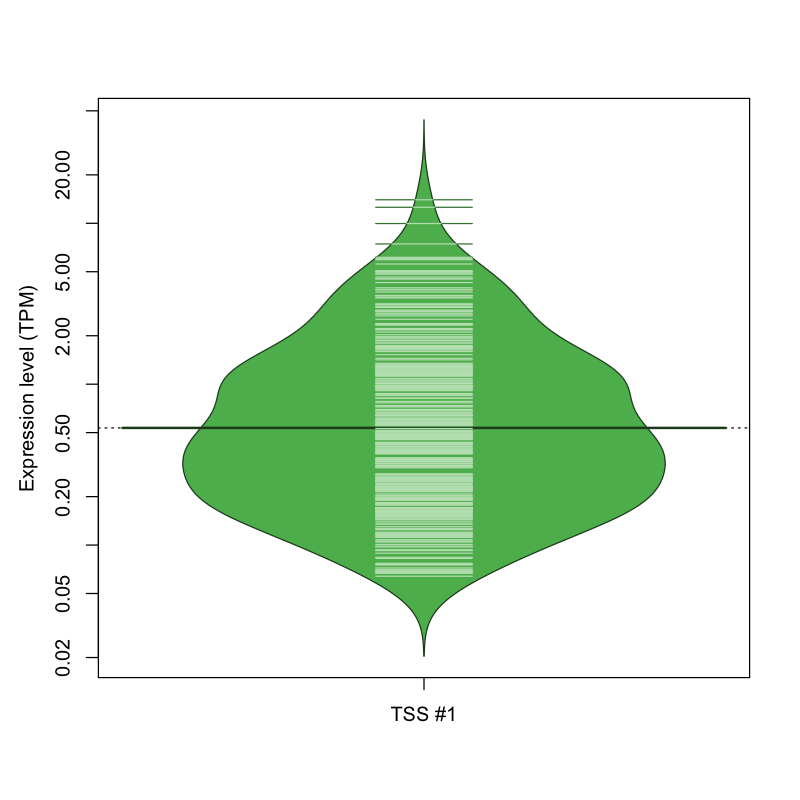
The graphical summary, for retro_mmus_1172 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1172 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1172 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 11 parental genes, and 17 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000018446 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000001993 | 5 retrocopies | |
| Dipodomys ordii | ENSDORG00000004514 | 1 retrocopy | |
| Felis catus | ENSFCAG00000022084 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000004937 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000033752 | 2 retrocopies |
retro_mmus_1172 , retro_mmus_3281,
|
| Nomascus leucogenys | ENSNLEG00000005936 | 1 retrocopy | |
| Oryctolagus cuniculus | ENSOCUG00000004594 | 2 retrocopies | |
| Ochotona princeps | ENSOPRG00000015738 | 1 retrocopy | |
| Sorex araneus | ENSSARG00000011001 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000004475 | 1 retrocopy |