RetrogeneDB ID: | retro_mmus_3267 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 9:40050589..40052716(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Rnf19a | ||
| Ensembl ID: | ENSMUSG00000022280 | ||
| Aliases: | Rnf19a, AA032313, Dorfin, Rnf19, UIP117, Ubce7ip2, XYbp | ||
| Description: | ring finger protein 19A [Source:MGI Symbol;Acc:MGI:1353623] |

Retrocopy-Parental alignment summary:
>retro_mmus_3267
GACTCTGACTCAGTTCTGATGAGCATTTTAGACATGAGTCTACATCAGCAAATGGGTTCAGAGGGAGATCGTCAACCTTC
TGCTTCCTCTGTGAGCTTGCCTTCTGTCAAAAATGCACCCAAACAGAGAAGATTTTCAATAGGCTCTCGTTTTTGGAGGA
AGATGGACAGCAAACGAAAATCAAGGGAGCTCGATGGTGGGGTGGAAGCAATTGCAAGTATTGAGAGGATACATTCTGAA
ATGTGTGCTGATAAGAACTCCATTTTCTCAACAAATACCTCTTCTGACAATGGATTGACTTCCATCAGCAAACAAACTGG
AGACTTTATGGACTGTCCTTTGTGCCTTTTGCGGCTTTCTCAAGACAGATTTCCTGATATCATGACATGTCATCATAGAT
CTTGTGTGGATTGTTTACGACAATACCTAAAGATAGAAATCTCTGAAAGCAGAATTAACATCAGTTGTCCAGAATGCCCT
GAACGGTTTAACCCCCATGACATTCGCTTGATATTAAGTGATGATGTCTTAATGGGAAAATATGAAGAATTTATGCTTCG
ACAGTGGCTTGTTGCAGATCCTGACTGTAGGTGGTGTCCGGCTCCAGACTGTGGATACGCTGTGATAGCATTTGGATGTG
CCAGGTGTCCAAAATTAACTTGTGGGCGAAAAGGCTGTGGAACAAAGTTTTGCTACCACTGTAAACAGATCTGGCATCCC
AAACAGACCTGTGAGACTGCTCTATGGGAAAGAGCCCAGCGTTTATGCTTAAGAACTTTACCCTTTTCACCTATTAGTTA
CAAACAGGAATCTAGAGCAGCAGCTGATGACATAAAGCCGTGTCCACGATGTGCTGCTTATATAATCAAGGTTGAGGATG
GGAGTTGCAATCATATGACATGTGCAGTCTGTGGTTGTGAGTTTTGTTGGTTGTGCATGAAAGAAATCTCCAATTTACAT
TATCTCAGTCCCTCGGGTTGTAGTTTATGGGGGAAGAAACCCTGGAGCCGAAAGAAGAAAATCCTGTGGCAGCTGGGAAC
CCTTGTTGGTGCTCCCGTCGGGATTGCTCTCATAGCCGGCATTGCCCTTCCTGCCATGATCATTGGCATTCCTGTGTATG
TGGGCCTCAAGATTCATGATTGCTATGAAGGCAGAGATGTTTCAAAACACAAGCGGAATCTGGTCATAGCAAGTGGTGTT
ACCCTGTCTGTAATCGTCTCTCCAGTGGTAGCTGCAGTGACTATAGGCATTGGTGTTCCTACCATGCTAGCTTATGTCTA
TGGTGTGGTCCCAATTTCCCTCTGCAGAAACAGGGATTGTGGAGAGTCTGCAGGAAATGGGAAAGGAGTAAGGATTGAGT
TTGATGATGAGAATGACATAAACGTAGGCGGGACTAACACAGCCATGGATACAACATCAGTAGCAGAAGCAAGGCACAAC
CCTAGCATAGGAGAGGGAAGTGTCAGCGGCCTGACTGGCAGTTTGAGTGCAAGTGGAAGCCACATGGATGGAATAGGCAC
TATTGGCAACAACCTGAGTGAGACAGTCAGTACCATAAGCTTAGCTGGAGCTAGTATCACAGGCAGTCTGTCAGGAAGTG
CCATGGTAAACTGTTTCAGCAATTTGCAAGTTCAAGCAGAGGTACAGAAAGAAAGGTGTAGTCTCAGTAGAGAATCTGGC
ACAGTGAGTTTGGGAAGAGTTTGTAATAACAGCAGCACCAAAGCAATGGCAGGATCCATTCTGAACTCCTACATCCCTCT
GAACAAAGATGGGAACAGTATGGAAGTACAAGTGGATATTGAATCAAAGTCATTCAAGTTCCAACACAGCCGTGGAAGCA
GCAGTGTGGATGACAGTGGTGCTGCCCGAGGCCACATGGGCAGGTCATCCAGTGGCTTGCCTGAAGGTAAATCCAGTGCT
ACCAAGTATTCCAAGGGAGCAACAGGAGGAAAAAAGTCAAAGAGTGGTAAACTGAGGAAAAAGGGTAACATGAAGATCAA
TGAGACCAGAGAGCACATGGATGCACAGTTGTTACAGCCTCAAAGCACAGATTTAAGTGAGTTTGAGGTGCCATTCCCGA
GTGGCAACTTGCCATCTGTAGCAGTCTCCCCCTGCAGTCACTCTTCT
ORF - retro_mmus_3267 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 85.19 % |
| Parental protein coverage: | 84.4 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | DSDSILMSILDMSLHQQMGSDRDLQSSTSSVSLPSVKKAPKQRRISIGSLFRRKKDSKRKSRELNGGVDG |
| DSDS.LMSILDMSLHQQMGS..D.Q.S.SSVSLPSVK.APKQRR.SIGS.F.RK.DSKRKSREL.GGV.. | |
| Retrocopy | DSDSVLMSILDMSLHQQMGSEGDRQPSASSVSLPSVKNAPKQRRFSIGSRFWRKMDSKRKSRELDGGVEA |
| Parental | IASIESIHSEMCADKNSIFSTNTSSDNGLTSISKQIGDFIECPLCLLRHSKDRFPDIMTCHHRSCVDCLR |
| IASIE.IHSEMCADKNSIFSTNTSSDNGLTSISKQ.GDF..CPLCLLR.S.DRFPDIMTCHHRSCVDCLR | |
| Retrocopy | IASIERIHSEMCADKNSIFSTNTSSDNGLTSISKQTGDFMDCPLCLLRLSQDRFPDIMTCHHRSCVDCLR |
| Parental | QYLRIEISESRVNISCPECTERFNPHDIRLILSDDVLMEKYEEFMLRRWLVADPDCRWCPAPDCGYAVIA |
| QYL.IEISESR.NISCPEC.ERFNPHDIRLILSDDVLM.KYEEFMLR.WLVADPDCRWCPAPDCGYAVIA | |
| Retrocopy | QYLKIEISESRINISCPECPERFNPHDIRLILSDDVLMGKYEEFMLRQWLVADPDCRWCPAPDCGYAVIA |
| Parental | FGCASCPKLTCGREGCGTEFCYHCKQIWHPNQTCDAARQERAQSLRLRTIRSSSISYSQESGAAADDIKP |
| FGCA.CPKLTCGR.GCGT.FCYHCKQIWHP.QTC..A..ERAQ.L.LRT...S.ISY.QES.AAADDIKP | |
| Retrocopy | FGCARCPKLTCGRKGCGTKFCYHCKQIWHPKQTCETALWERAQRLCLRTLPFSPISYKQESRAAADDIKP |
| Parental | CPRCAAYIIKMNDGSCNHMTCAVCGCEFCWLCMKEISDLHYLSPSGCTFWGKKPWSRKKKILWQLGTLVG |
| CPRCAAYIIK..DGSCNHMTCAVCGCEFCWLCMKEIS.LHYLSPSGC..WGKKPWSRKKKILWQLGTLVG | |
| Retrocopy | CPRCAAYIIKVEDGSCNHMTCAVCGCEFCWLCMKEISNLHYLSPSGCSLWGKKPWSRKKKILWQLGTLVG |
| Parental | APVGIALIAGIAIPAMIIGIPVYVGRKIHNRYEGKDVSKHKRNLAIAGGVTLSVIVSPVVAAVTVGIGVP |
| APVGIALIAGIA.PAMIIGIPVYVG.KIH..YEG.DVSKHKRNL.IA.GVTLSVIVSPVVAAVT.GIGVP | |
| Retrocopy | APVGIALIAGIALPAMIIGIPVYVGLKIHDCYEGRDVSKHKRNLVIASGVTLSVIVSPVVAAVTIGIGVP |
| Parental | IMLAYVYGVVPISLCRSGGCGVSAGNGKGVRIEFDDENDINVGGTNAAIDTTSVAEARHNPSIGEGSVGG |
| .MLAYVYGVVPISLCR...CG.SAGNGKGVRIEFDDENDINVGGTN.A.DTTSVAEARHNPSIGEGSV.G | |
| Retrocopy | TMLAYVYGVVPISLCRNRDCGESAGNGKGVRIEFDDENDINVGGTNTAMDTTSVAEARHNPSIGEGSVSG |
| Parental | LTGSLSASGSHMDRIGTIRDNLSETASTMALAGASITGSLSGSAMVNCFNRLEVQADVQKERCSLSGESG |
| LTGSLSASGSHMD.IGTI..NLSET.ST..LAGASITGSLSGSAMVNCF..L.VQA.VQKERCSLS.ESG | |
| Retrocopy | LTGSLSASGSHMDGIGTIGNNLSETVSTISLAGASITGSLSGSAMVNCFSNLQVQAEVQKERCSLSRESG |
| Parental | TVSLGTVSDNASTKAMAGSILNSYIPLDREGNSMEVQVDIESKPFKFRHNSGSSSVDDSGATRGHTGGAS |
| TVSLG.V..N.STKAMAGSILNSYIPL...GNSMEVQVDIESK.FKF.H..GSSSVDDSGA.RGH.G..S | |
| Retrocopy | TVSLGRVCNNSSTKAMAGSILNSYIPLNKDGNSMEVQVDIESKSFKFQHSRGSSSVDDSGAARGHMGRSS |
| Parental | SGLPEGKSSATKWSKEATGGKKSKSGKLRKKGNMKINETREDMDAQLLEQQSTNSSEFEAPSLSDSMPSV |
| SGLPEGKSSATK.SK.ATGGKKSKSGKLRKKGNMKINETRE.MDAQLL..QST..SEFE.P..S...PSV | |
| Retrocopy | SGLPEGKSSATKYSKGATGGKKSKSGKLRKKGNMKINETREHMDAQLLQPQSTDLSEFEVPFPSGNLPSV |
| Parental | ADSHSSHFS |
| A.S..SH.S | |
| Retrocopy | AVSPCSHSS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .05 RPM | 38 .95 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 36 .82 RPM |
| SRP007412_heart | 0 .00 RPM | 24 .16 RPM |
| SRP007412_kidney | 0 .00 RPM | 51 .78 RPM |
| SRP007412_liver | 0 .14 RPM | 36 .89 RPM |
| SRP007412_testis | 373 .00 RPM | 554 .72 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_3267 was not detected
1 EST(s) were mapped to retro_mmus_3267 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA064465 | 40052325 | 40052437 | 97.3 | 111 | 0 | 109 |
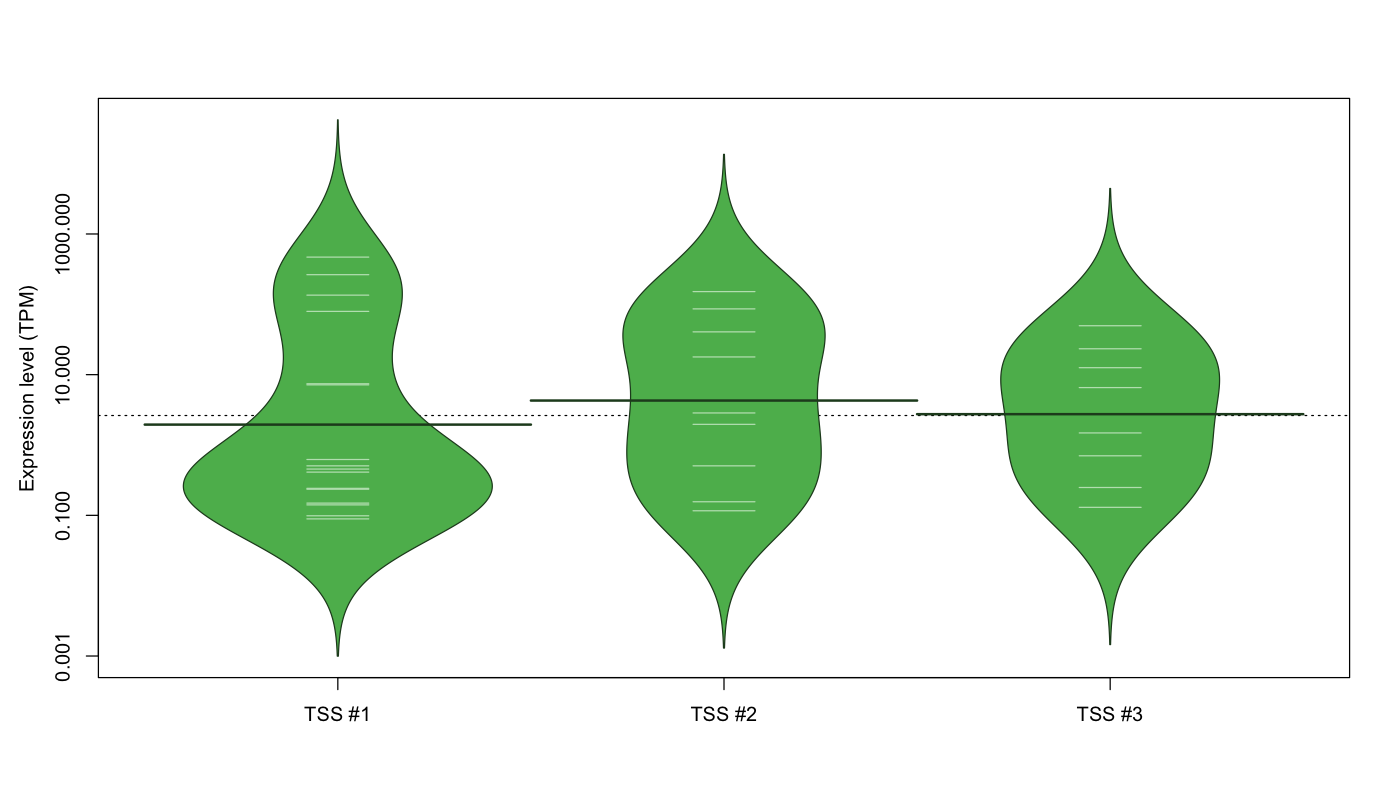
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_149642 | 1056 libraries | 10 libraries | 0 libraries | 2 libraries | 4 libraries |
| TSS #2 | TSS_149643 | 1063 libraries | 3 libraries | 2 libraries | 0 libraries | 4 libraries |
| TSS #3 | TSS_149644 | 1064 libraries | 3 libraries | 1 library | 1 library | 3 libraries |
The graphical summary, for retro_mmus_3267 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3267 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3267 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 1 parental gene, and 3 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Mus musculus | ENSMUSG00000022280 | 3 retrocopies |
retro_mmus_3267 , retro_mmus_3275, retro_mmus_3277,
|