RetrogeneDB ID: | retro_mmus_68 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 11:91575448..91577455(-) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000046755 | |
| Aliases: | Kif2b, 1700063D03Rik | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Kif2a | ||
| Ensembl ID: | ENSMUSG00000021693 | ||
| Aliases: | Kif2a, C530030B14Rik, Kif2, Kns2, M-kinesin | ||
| Description: | kinesin family member 2A [Source:MGI Symbol;Acc:MGI:108390] |

Retrocopy-Parental alignment summary:
>retro_mmus_68
ATGGCCAGCCAGTTCTGCCTTCCTCTGGCCCCACGCCTCTCACCCCTTAAACCTCTGAAATCCCACTTCACAGACTTCCA
GGTTGGCATCTGTGTGGCAATCCAGCGCAGTGACAAACGGATTCACCTGGCTGTGGTCACAGAGATCAACAGAGAAAACT
CCTGGGTCACAGTAGAGTGGGTTGAGAAAGGAGTCAAAAAAGGAAAGAAGATCGAACTGGAGACTGTACTTCTGTTGAAT
CCGGCTCTGGCCTCTCTTGAACACCAACGGTCACGTAGGCCCCTGCGCCCAGTGTCTGTAGTACCATCTACTGCCATTGG
AGACCAGCGTACAGCCACCAAGTGGATTGCGATGATCCCACACAGAAATGAAACACCCTCAGGGGACAGCCAAACCCTGA
TGATCCCCAGCAACCCTTGTCTGATGAAGCGGAAAAAATCCCCCTGCCTACGGGAAATTGAAAAGCTGCAGAAGCAGAGA
GAGAAGCGCCGCCGACTGCAGCTAGAGATCCGAGCTAGACGAGCCCTTGACATCAACACTGGAAATCCGAACTTCGAGAC
TATGCGCATGATTGAGGAGTATCGCAGGCGTCTAGATAGCAGCAAGATGTCCAGCCTGGAGCCCCCAGAAGACCACCGGA
TCTGTGTGTGTGTGAGAAAGCGCCCTCTTAATCAGAGAGAGACGACGATGAAGGACCTCGATATCATCACTATTCCCTCG
CACAATGTAGTCATGGTCCACGAGTCTAAACAAAAGGTGGACCTGACCCGCTACTTGGAAAACCAGACCTTCTGTTTTGA
CCACGCCTTCGACGACAAGGCATCCAATGAACTAGTTTACCAGTTCACCGCCAGGCCTCTTGTGGAGTCCATCTTCCGAA
AGGGCATGGCAACTTGCTTTGCCTACGGGCAGACTGGCAGTGGAAAAACCCACACGATGGGCGGAGCCTTTTTAGGAAAG
GCCCAAGATTGTTCTAAAGGCATTTACGCCCTGGTAGCTCAGGATGTCTTCCTCCTACTAAAAACCCCAGCCTATGAGAA
ATTGGAACTCAAAGTCTATGGGACCTTTTTTGAGATCTATGGTGGCAAAGTGTACGATCTGTTGAACTGGAAGAAGAAAC
TACAAGTCCTGGAGGATGGAAATCAGCAAGTCCAAGTGGTGGGGCTGCAGGAGCAGGAGGTGAGCTGTGTGGAGGAAGTC
CTGAATCTGGTGGAGCTCGGGAACAGCTGTCGGACTTCCGGTCAGACCTCGGTCAACGCCCACTCCTCCAGGAGCCATGC
TGTGTTCCAGCTCATTCTCAAGGCTGGAGGGAAGCTGCACGGCAAGTTTTCCCTGGTTGACTTAGCTGGGAACGAAAGGG
GAGCAGATACTGCCAAGGCCACCCGGAAGAGGCAGCTGGAAGGGGCTGAGATCAATAAAAGTCTGCTAGCCCTGAAAGAG
TGCATCAGGGCTTTGGGTAAGAACAAGTCTCATACCCCATTCAGAGCCAGCAAACTCACGCAGGTGCTACGGGATTCCTT
CATAGGGCAGAACTCCTATACGTGCATGATCGCGACAATCTCACCCGGGATGACTTCCTGTGAGAATACTCTCAATACTT
TAAGATACGCCAACAGAGTCAAAGAATTAGCTTTAGAGGCAAGGCCCTACCATCACTGTGTCTCTCCACCTGGCCATGAG
GTACCACTGATGATAGAAAACGACAATACGAATTCGGGAAAGTCCCTTCAGAGGGATGAAGTTATTCAGATACCTACTGT
AGAGAAAGAGGAAGAGAAAGAAAGTGATGAACTCACATCAAAAAAGGAGCCAGCAGCTTCATGGAGCAGGTCCAACCAGT
GGTGGGAGGCCATCCAAGAAACGGCTGAGGGAGTCAACGGTGATGTGGATTTTTGCATTGCGCAGTCTTTGTCTGTTTTG
GAGCAGAAAATCGGTGTGCTGACTGATATCCAAAAAAAGCTCCAATCACTACGAGAAGACCTCCAAAAGAAAAGCCAAGT
TGAGTGA
ORF - retro_mmus_68 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 76.12 % |
| Parental protein coverage: | 57.02 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | RRKSNCVKEVEKLQEKREKRRLQQQELREKRAQDVDATNPNYEIMCMIRDFRGSLDYRPLTTADPIDEHR |
| R.KS.C..E.EKLQ..REKRR..Q.E.R..RA.D....NPN.E.M.MI...R..LD........P...HR | |
| Retrocopy | RKKSPCLREIEKLQKQREKRRRLQLEIRARRALDINTGNPNFETMRMIEEYRRRLDSSKMSSLEPPEDHR |
| Parental | ICVCVRKRPLNKKETQMKDLDVITIPSKDVVMVHEPKQKVDLTRYLENQTFRFDYAFDDSAPNEMVYRFT |
| ICVCVRKRPLN..ET.MKDLD.ITIPS..VVMVHE.KQKVDLTRYLENQTF.FD.AFDD.A.NE.VY.FT | |
| Retrocopy | ICVCVRKRPLNQRETTMKDLDIITIPSHNVVMVHESKQKVDLTRYLENQTFCFDHAFDDKASNELVYQFT |
| Parental | ARPLVETIFERGMATCFAYGQTGSGKTHTMGGDFSGKNQDCSKGIYALAARDVFLMLKKPNYKKLELQVY |
| ARPLVE.IF..GMATCFAYGQTGSGKTHTMGG.F.GK.QDCSKGIYAL.A.DVFL.LK.P.Y.KLEL.VY | |
| Retrocopy | ARPLVESIFRKGMATCFAYGQTGSGKTHTMGGAFLGKAQDCSKGIYALVAQDVFLLLKTPAYEKLELKVY |
| Parental | ATFFEIYSGKVFDLLNRKTKLRVLEDGKQQVQVVGLQEREVKCVEDVLKLIDIGNSCRTSGQTSANAHSS |
| .TFFEIY.GKV.DLLN.K.KL.VLEDG.QQVQVVGLQE.EV.CVE.VL.L...GNSCRTSGQTS.NAHSS | |
| Retrocopy | GTFFEIYGGKVYDLLNWKKKLQVLEDGNQQVQVVGLQEQEVSCVEEVLNLVELGNSCRTSGQTSVNAHSS |
| Parental | RSHAVFQIILRRKGKLHGKFSLIDLAGNERGADTSSADRQTRLEGAEINKSLLALKECIRALGRNKPHTP |
| RSHAVFQ.IL...GKLHGKFSL.DLAGNERGADT..A.R...LEGAEINKSLLALKECIRALG.NK.HTP | |
| Retrocopy | RSHAVFQLILKAGGKLHGKFSLVDLAGNERGADTAKATRKRQLEGAEINKSLLALKECIRALGKNKSHTP |
| Parental | FRASKLTQVLRDSFIGENSRTCMIATISPGMASCENTLNTLRYANRVKELTV |
| FRASKLTQVLRDSFIG.NS.TCMIATISPGM.SCENTLNTLRYANRVKEL.. | |
| Retrocopy | FRASKLTQVLRDSFIGQNSYTCMIATISPGMTSCENTLNTLRYANRVKELAL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .07 RPM | 86 .07 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 56 .71 RPM |
| SRP007412_heart | 0 .00 RPM | 20 .17 RPM |
| SRP007412_kidney | 0 .00 RPM | 20 .14 RPM |
| SRP007412_liver | 0 .08 RPM | 9 .28 RPM |
| SRP007412_testis | 603 .07 RPM | 244 .29 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_68 was not detected
No EST(s) were mapped for retro_mmus_68 retrocopy.
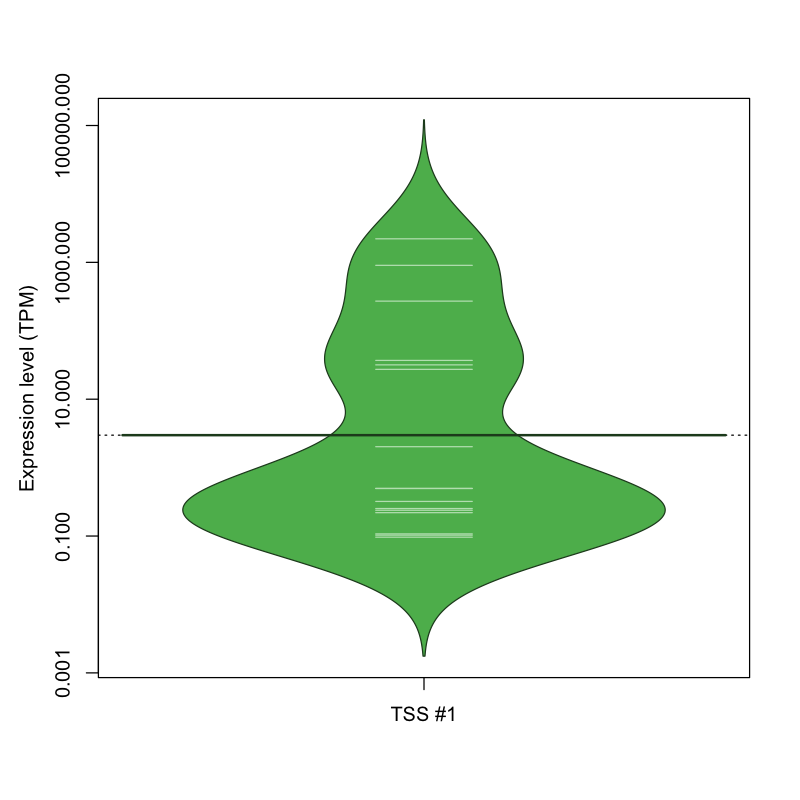
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_18859 | 1056 libraries | 9 libraries | 1 library | 0 libraries | 6 libraries |
The graphical summary, for retro_mmus_68 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_68 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_68 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_47 |