RetrogeneDB ID: | retro_mmus_297 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:14865071..14865962(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000082193 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Rpl5 | ||
| Ensembl ID: | ENSMUSG00000058558 | ||
| Aliases: | Rpl5, U21RNA | ||
| Description: | ribosomal protein L5 [Source:MGI Symbol;Acc:MGI:102854] |

Retrocopy-Parental alignment summary:
>retro_mmus_297
ATGGGGTTTGTGAAAGTTATCAAGAATAAGGCCTACTTTAAAAGATACCAAGTGAGATTTCAAAGGCGGCGAGAGGGTAA
AACTGACTACTATGCTCGAAAACGATTGGTGATCCAGGACAAGAATAAGTACAACACACCCAAATATAGGATGATAGTTC
GTGTAACTAACAGAGATATCATCTGCCAGATTGCATATGCCCGTATAGAAGGGGATATGATCGTCTGTGCAGCATATGCA
CATGAACTTCCAAAATATGGTGTGAAAGTTGGCCTGACAAATTATGCTGCAGCCTATTGCACTGGCCTGCTGCTGGCCCG
CAGGCTTCTGAATAGGTTTGGCATGGACAAGATCTATGAAGGCCAAGTGGAGGTGAATGGAGGTGAATACAATGTGGAAA
GCATTGACGGTCAGCCTGGTGCCTTCACTTGCTATCTGGATGCAGGTCTTGCCCGAACTACAACTGGCAATAAAGTTTTT
GGGGCCCTGAAGGGAGCTGTGGATGGAGGCTTGTCTATCCCTCATAGTACTAAACGATTCCCTGGTTATGACTCTGAAAG
CAAGGTGTTCAATGCAGAGGTACATCGGAAGCACATCATGGGTCAAAATGTGGCAGACTACATGTGCTACCTAATGGAGG
AAGATGAAGATGCGTATAAGAAACAGTTCTCTCAGTACATCAAGAACAATGTAACTCCAGACAAGATGGAGGAGATGTAT
AAGAAAGCTCATGCTGCTATCCGAGAGAATCCAGTCTATGAGAAGAAGCCCAAGAGAGAAGTGAAGAAGAAAAGGTGGAA
TCGTCCCAAAATGTCTCTTGCCCAGAAGAAAGACCGGGTTGCTCAAAAGAAGGCAAGCTTCCTCAGAGCTCAGGAAAGGG
CTGCTGAAAGC
ORF - retro_mmus_297 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 98.32 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MGFVKVVKNKAYFKRYQVRFRRRREGKTDYYARKRLVIQDKNKYNTPKYRMIVRVTNRDIICQIAYARIE |
| MGFVKV.KNKAYFKRYQVRF.RRREGKTDYYARKRLVIQDKNKYNTPKYRMIVRVTNRDIICQIAYARIE | |
| Retrocopy | MGFVKVIKNKAYFKRYQVRFQRRREGKTDYYARKRLVIQDKNKYNTPKYRMIVRVTNRDIICQIAYARIE |
| Parental | GDMIVCAAYAHELPKYGVKVGLTNYAAAYCTGLLLARRLLNRFGMDKIYEGQVEVNGGEYNVESIDGQPG |
| GDMIVCAAYAHELPKYGVKVGLTNYAAAYCTGLLLARRLLNRFGMDKIYEGQVEVNGGEYNVESIDGQPG | |
| Retrocopy | GDMIVCAAYAHELPKYGVKVGLTNYAAAYCTGLLLARRLLNRFGMDKIYEGQVEVNGGEYNVESIDGQPG |
| Parental | AFTCYLDAGLARTTTGNKVFGALKGAVDGGLSIPHSTKRFPGYDSESKEFNAEVHRKHIMGQNVADYMRY |
| AFTCYLDAGLARTTTGNKVFGALKGAVDGGLSIPHSTKRFPGYDSESK.FNAEVHRKHIMGQNVADYM.Y | |
| Retrocopy | AFTCYLDAGLARTTTGNKVFGALKGAVDGGLSIPHSTKRFPGYDSESKVFNAEVHRKHIMGQNVADYMCY |
| Parental | LMEEDEDAYKKQFSQYIKNNVTPDMMEEMYKKAHAAIRENPVYEKKPKREVKKKRWNRPKMSLAQKKDRV |
| LMEEDEDAYKKQFSQYIKNNVTPD.MEEMYKKAHAAIRENPVYEKKPKREVKKKRWNRPKMSLAQKKDRV | |
| Retrocopy | LMEEDEDAYKKQFSQYIKNNVTPDKMEEMYKKAHAAIRENPVYEKKPKREVKKKRWNRPKMSLAQKKDRV |
| Parental | AQKKASFLRAQERAAES |
| AQKKASFLRAQERAAES | |
| Retrocopy | AQKKASFLRAQERAAES |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .12 RPM | 46 .80 RPM |
| SRP007412_cerebellum | 0 .13 RPM | 34 .26 RPM |
| SRP007412_heart | 0 .12 RPM | 57 .28 RPM |
| SRP007412_kidney | 0 .06 RPM | 78 .35 RPM |
| SRP007412_liver | 0 .11 RPM | 59 .15 RPM |
| SRP007412_testis | 0 .14 RPM | 29 .04 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_297 was not detected
No EST(s) were mapped for retro_mmus_297 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_70063 | 130 libraries | 249 libraries | 456 libraries | 159 libraries | 78 libraries |
The graphical summary, for retro_mmus_297 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
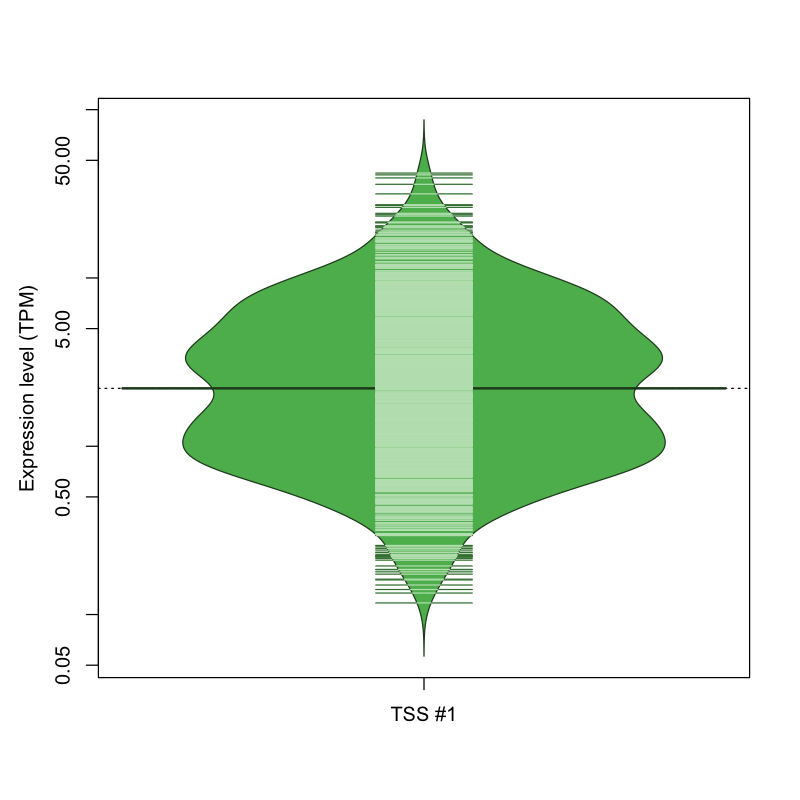
retro_mmus_297 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_297 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 16 parental genes, and 56 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000009878 | 5 retrocopies | |
| Bos taurus | ENSBTAG00000002026 | 2 retrocopies | |
| Equus caballus | ENSECAG00000021065 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000006361 | 3 retrocopies | |
| Monodelphis domestica | ENSMODG00000011635 | 1 retrocopy | |
| Mustela putorius furo | ENSMPUG00000004150 | 6 retrocopies | |
| Mus musculus | ENSMUSG00000058558 | 11 retrocopies | |
| Ornithorhynchus anatinus | ENSOANG00000004669 | 1 retrocopy | |
| Oryctolagus cuniculus | ENSOCUG00000010089 | 2 retrocopies | |
| Petromyzon marinus | ENSPMAG00000002040 | 1 retrocopy | |
| Pteropus vampyrus | ENSPVAG00000015117 | 4 retrocopies | |
| Rattus norvegicus | ENSRNOG00000023529 | 6 retrocopies | |
| Sarcophilus harrisii | ENSSHAG00000006417 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000006899 | 2 retrocopies | |
| Tupaia belangeri | ENSTBEG00000010560 | 5 retrocopies | |
| Tursiops truncatus | ENSTTRG00000013097 | 5 retrocopies |